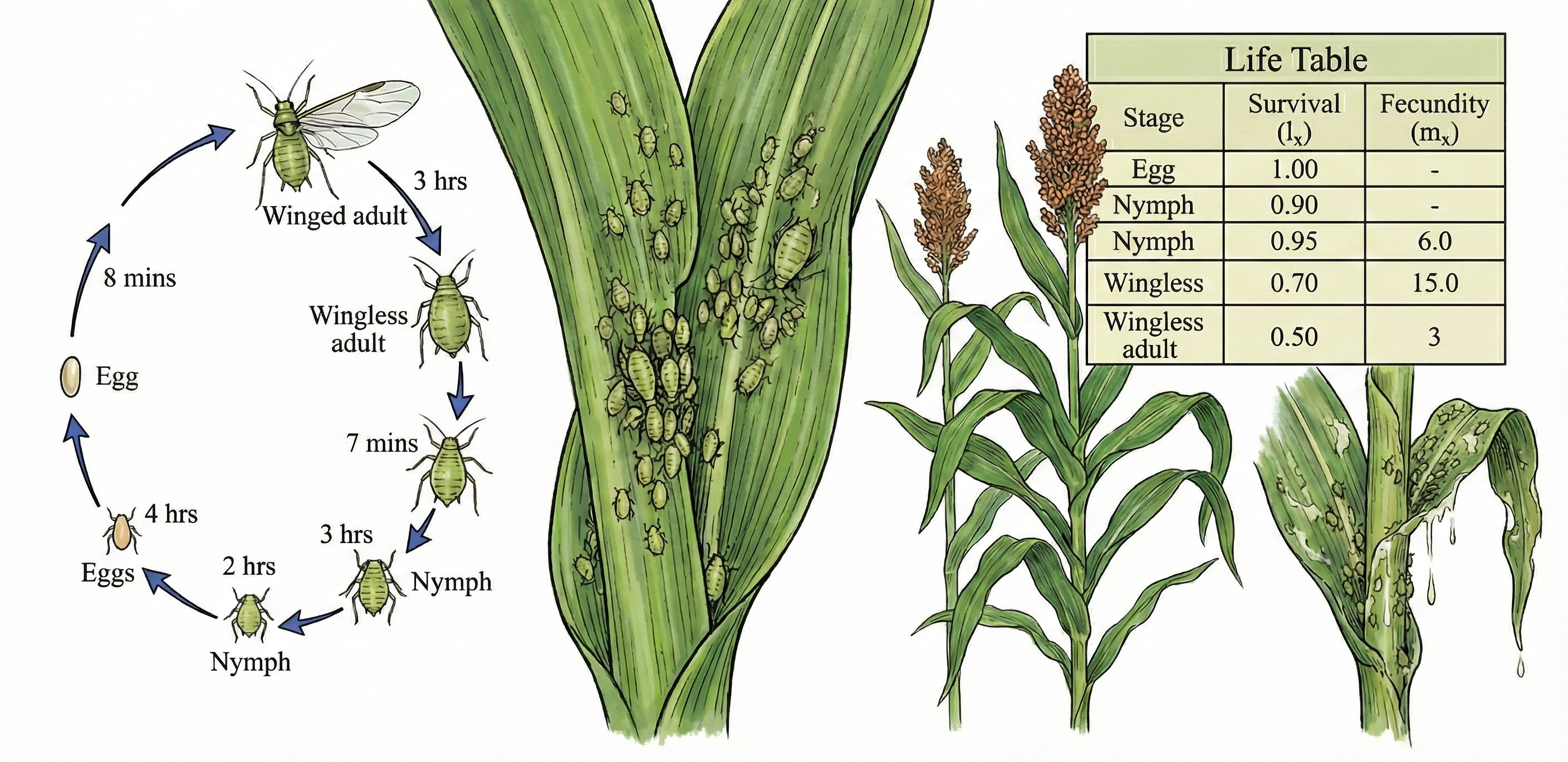
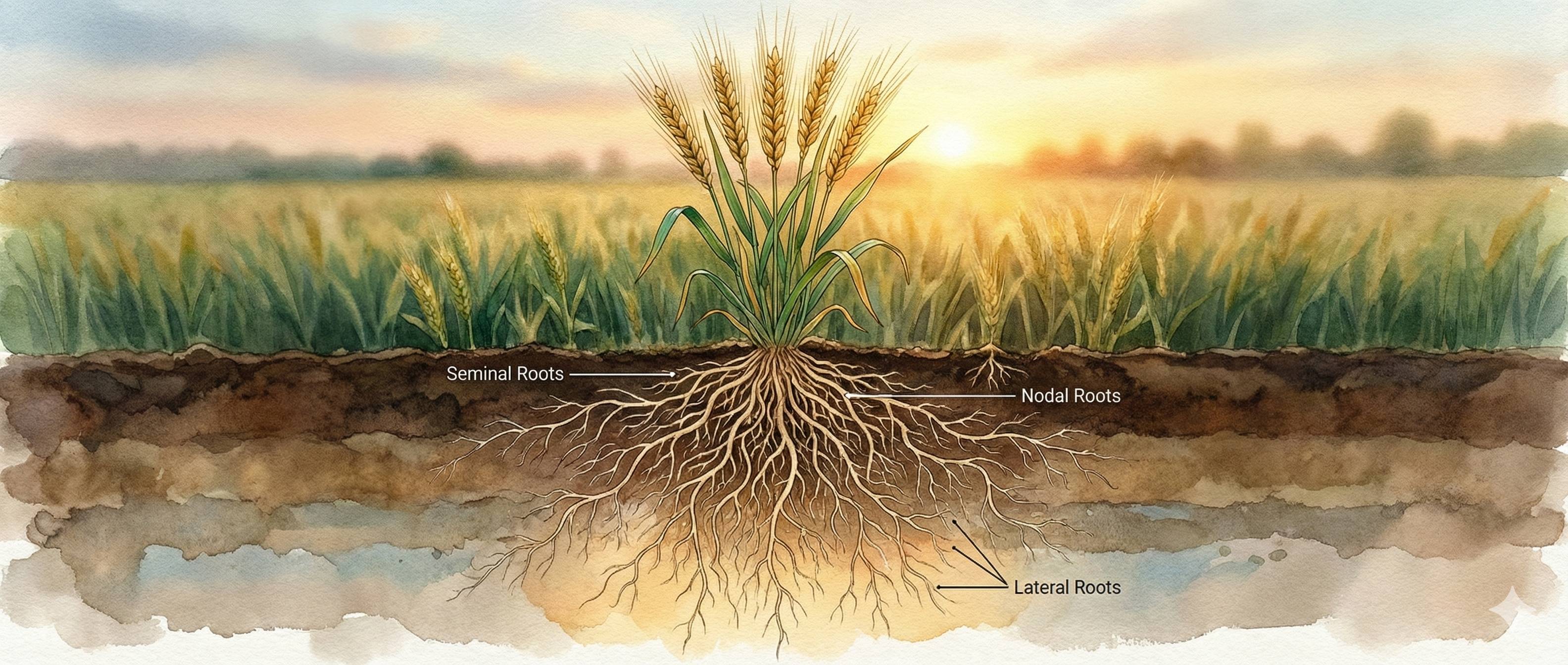
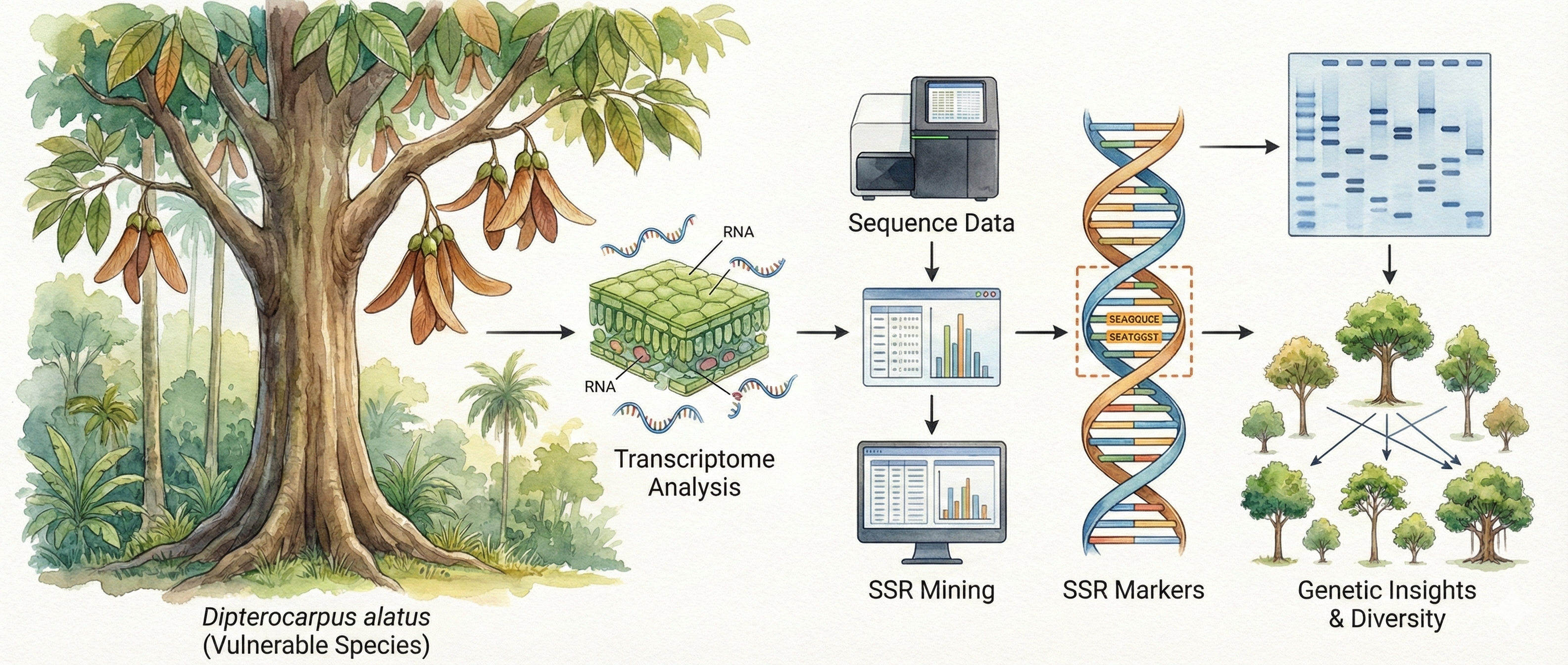
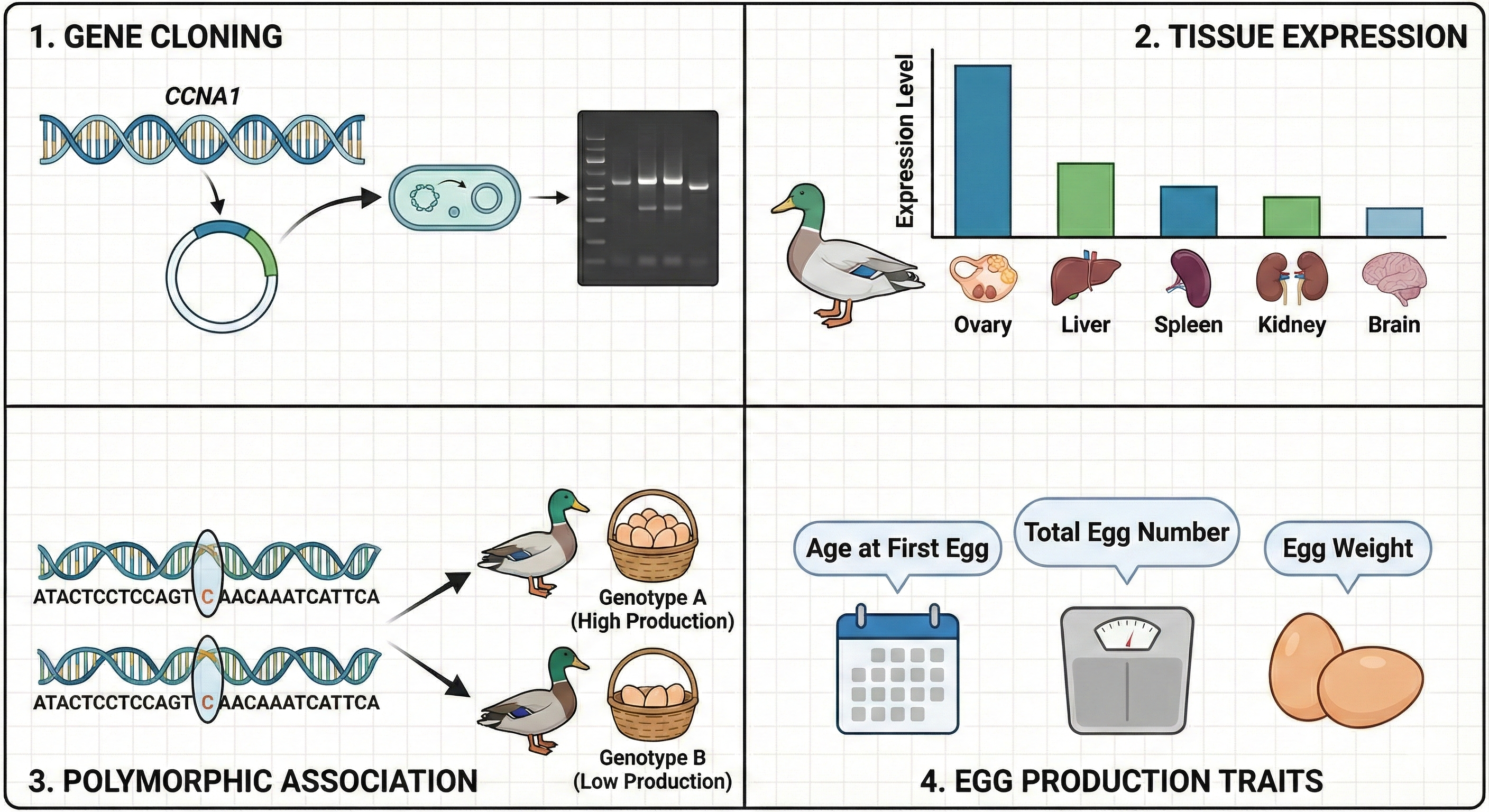
About the Journal
Genetics and Molecular Research (GMR) publishes high quality research in genetics and molecular biology. GMR reflects the full breadth and interdisciplinary nature of this research by publishing outstanding original contributions in all areas of biology.
GMR publishes human studies, as well as research on model organisms—from mice and flies, to plants and bacteria. Our emphasis is on studies of broad interest that provide significant insight into a biological process or processes. Topics include, but are not limited to gene discovery and function, population genetics, evolution, genome projects, comparative and functional genomics, molecular analysis of simple and complex genetic traits, cancer genetics, medical genetics, disease biology, agricultural genomics, developmental genetics, regulatory variation in gene expression, pharmacological genomics, evolution, gene expression, chromosome biology, and epigenetics.